Introduction
RAD is a webtool for analysing region associated differentially expressed genes. Visualization of DEGs surrounding peak regions are implemented in RAD in order to help researchers to infer the regulatory function of transcription factors or genomic sequence.
The algorithm behind RAD has been implemented in recent publications (Pastor et al., 2018; Harris et al., 2018; Gallego-Bartolomé et al., 2019). There are three main steps in the algorithm:
-
Infer region associated genes within specific distance using
awkandbedtools. -
A specific distance was split into different bins into which up/down-regulated genes are assigned. We calculated observed versus expected as follows to reduce false-positive rate:
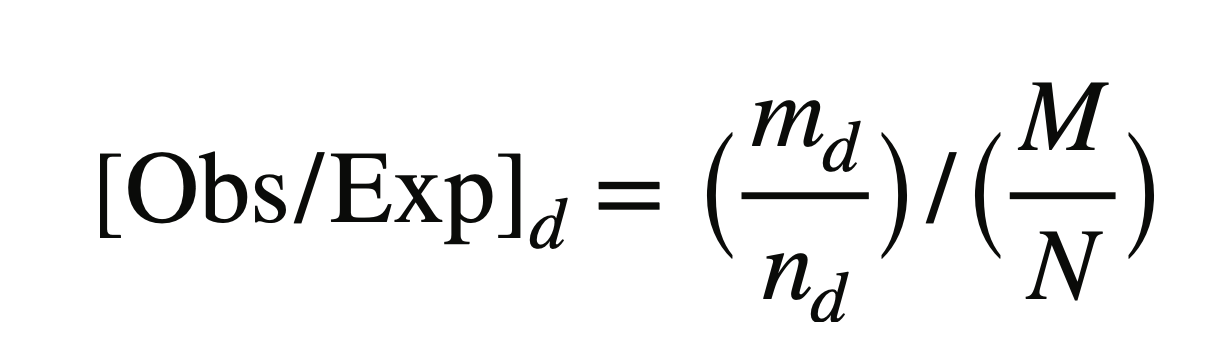
-
Perform a hypergeometric test to calculate the p-value over the four elements above
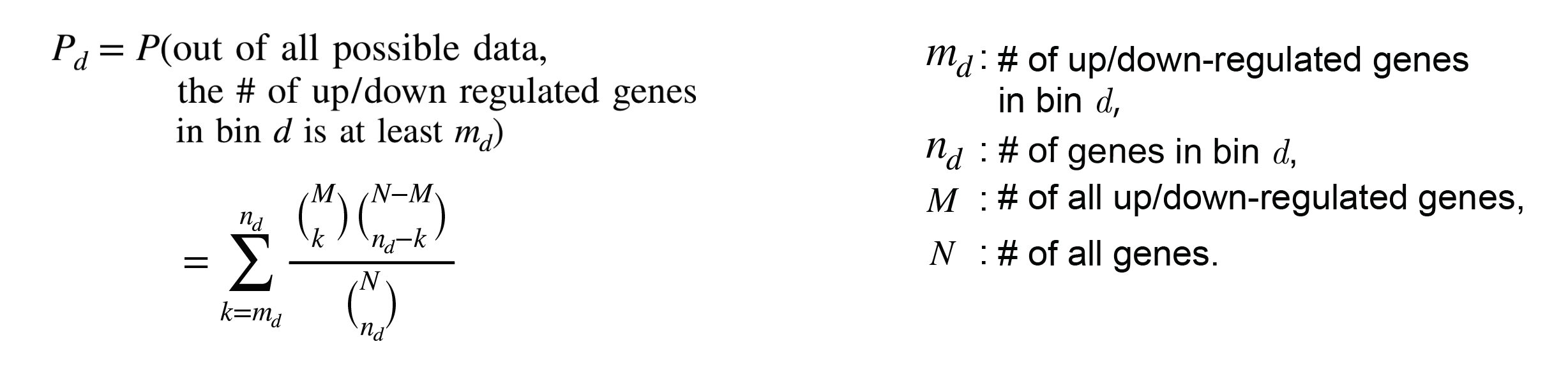
k, up/down-regulated genes covered in different distance bins,
n, all genes covered in different distance bins,
m, up/down-regulated genes covered in region extended distance,
N, whole genome genes.
Supported Reference Genome
Currently, RAD supports referenece genome of the well-annotated human (GRCh38/hg38 (EMSEMBL GRCh38, Dec. 2013) and GRCh37/hg19 (EMSEMBL GRCh37, Feb. 2009)), mouse (GRCm38/mm10 (ENSEMBL GRCm38, Dec. 2011) and NCBIm37 (NCBI build 37/mm9, Jul. 2007)), and Arabidopsis thaliana (EMSEMBL TAIR10, June. 2016).
We hope to gradually expand the set of genome data we support.
Terms and Conditions of Use
Disclaimer
The content of the pages of this website is for your general information and use only. It is subject to change without notice.
Neither we nor any third parties provide any warranty or guarantee as to the accuracy, timeliness, performance, completeness or suitability of the information and materials found or offered on this website for any particular purpose. You acknowledge that such information and materials may contain inaccuracies or errors and we expressly exclude liability for any such inaccuracies or errors to the fullest extent permitted by law.
Your use of any information or materials on this website is entirely at your own risk, for which we shall not be liable. It shall be your responsibility to ensure that any products, services or information available through this website meet your specific requirements.
This website contains material which is owned by or licensed to us. This material includes, but is not limited to, the design, layout, look, and appearance. Reproduction is prohibited other than under the copyright notice, which forms part of these terms and conditions. All trademarks reproduced in this website which is not the property of, or licensed to, the operator is acknowledged on the website.
All trade marks reproduced in this website which are not the property of, or licensed to, the operator are acknowledged on the website.
Permission and limitation of use
User may download material from the websites only for User’s own personal, non-commercial use. You may our website for legal use only.
Links to Other Sites
Our websites contain hyperlinks to other websites maintained or controlled by others. We do not have the responsibility to approve, review or endorse the content of any products and service that may be provided by these websites.
Citations
Here are some papers that cites RAD:
- Pastor, W.A., Liu, W., Chen, D., Ho, J., Kim, R., Hunt, T.J., Lukianchikov, A., Liu, X., Polo, J.M., Jacobsen, S.E. & Clark, A.T. (2018) TFAP2C regulates transcription in human naive pluripotency by opening enhancers. Nature cell biology, 20, 553-564. doi: 10.1038/s41556-018-0089-0
- Harris, C.J., Scheibe, M., Wongpalee, S.P., Liu, W., Cornett, E.M., Vaughan, R.M., Li, X., Chen, W., Xue, Y., Zhong, Z., Yen, L., Barshop, W.D., Rayatpisheh, S., Gallego-Bartolome, J., Groth, M., Wang, Z., Wohlschlegel, J.A., Du, J., Rothbart, S.B., Butter, F. & Jacobsen, S.E. (2018) A DNA methylation reader complex that enhances gene transcription. Science, 362, 1182-1186. doi: 10.1126/science.aar7854
- Gallego-Bartolomé, J., Liu, W., Kuo, P.H., Feng, S., Ghoshal, B., Gardiner, J., Zhao, J.M., Park, S.Y., Chory, J. & Jacobsen, S.E. (2019) Co-targeting RNA Polymerases IV and V Promotes Efficient De Novo DNA Methylation in Arabidopsis. Cell, 176, 1068-1082.e19. doi: 10.1016/j.cell.2019.01.029
Web Browsers
Which browser is recommended for RAD?
We recommended Google Chrome. It offers the best compatibility and performance in our tests.
On which browsers and platforms has RAD been tested?
RAD was tested on:
- Google Chrome on MacOS and Windows
- Safari on MacOS
About Us
Team Members
- Wanlu Liu (Principle Investigator) wanluliu@intl.zju.edu.cn
- Yixin Guo yixinguo.19@intl.zju.edu.cn
- Ziwei Xue ziwei.17@intl.zju.edu.cn
- Ruihong Yuan ruihong.19@intl.zju.edu.cn
Report an Issue
Feel free to report any issue or bug to us by sending email to yixinguo.19@intl.zju.edu.cn or ziwei.17@intl.zju.edu.cn