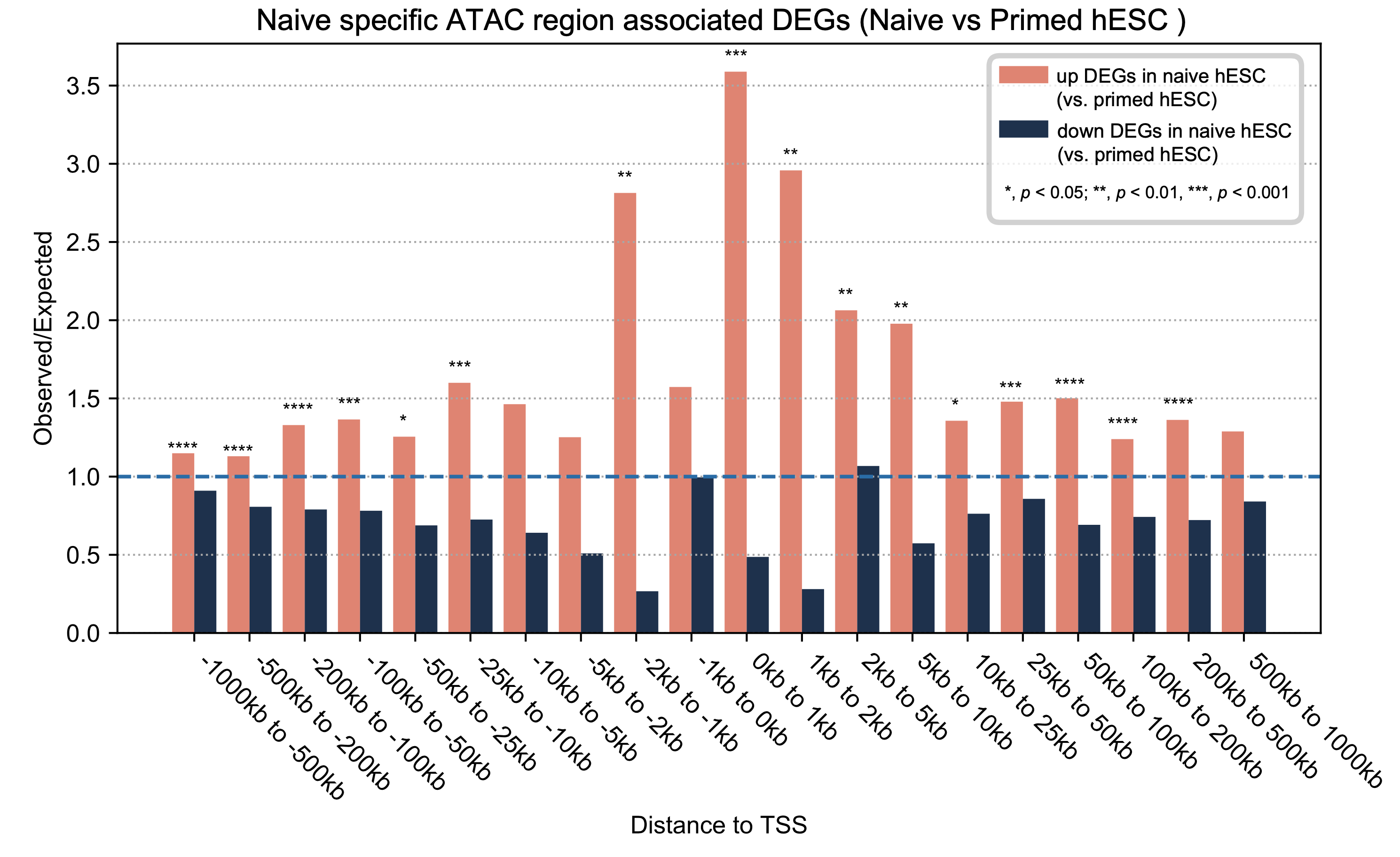
RAD - Region Associated DEG : Identify Region Associated Differentially Expressed Genes
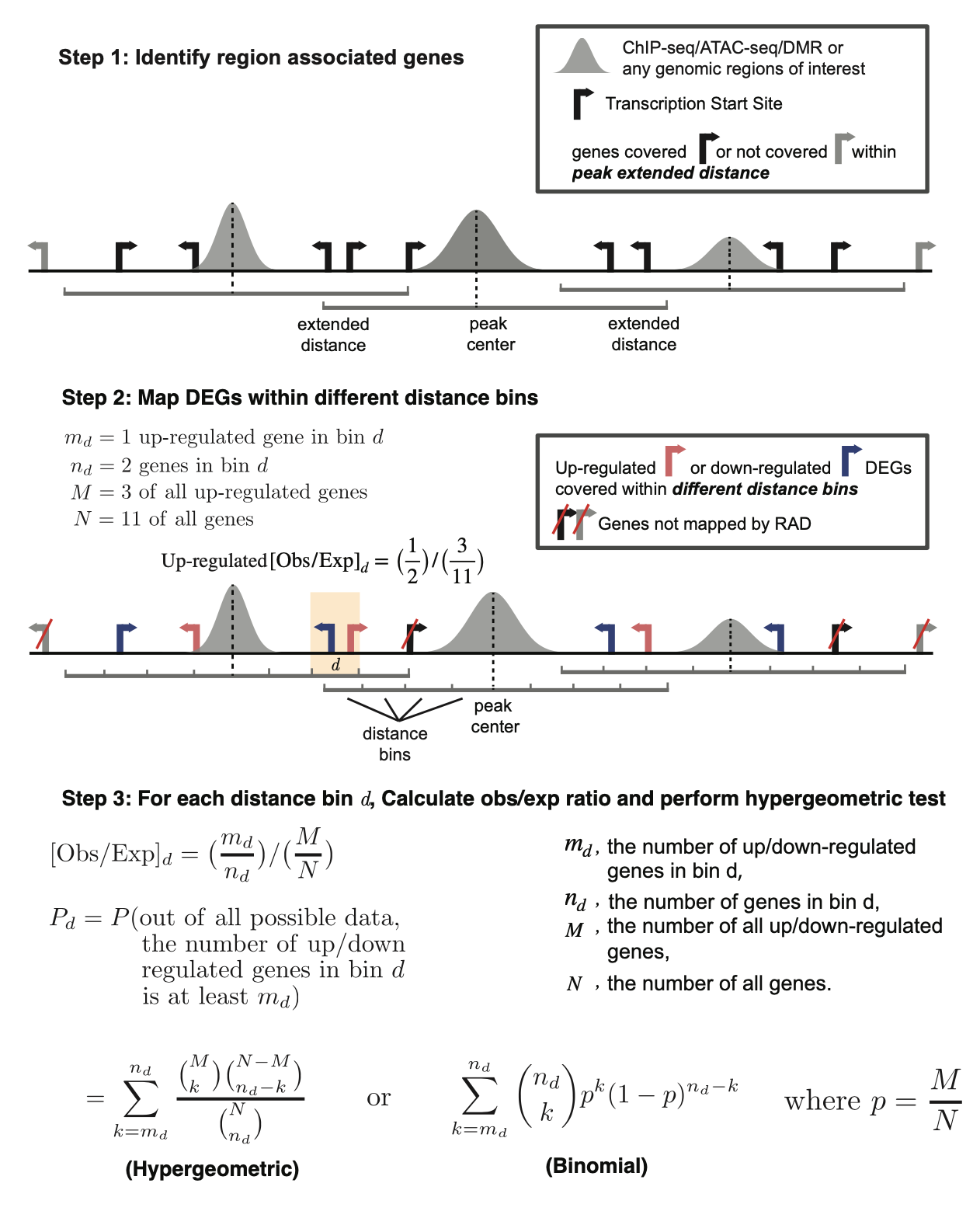
Region Associated DEG (RAD) is a tool to find region associated differentially expressed genes.
The algorithm behind RAD has been implemented in recent publications (Liu et al., Nat. Commun., 2021. ; Pastor, W.A. et al., Nat. Cell Biol., 2018; Harris, C.J. et al., Science, 2018; Gallego-Bartolomé, J. et al., Cell, 2019).
See supported format for upload in the example.
News
-
Feb. 2, 2021:RAD: a web application to identify region associated differentially expressed genes was published online on Bioinformatics.
-
Jan. 28, 2021:The associated paper "RAD: a web application to identify region associated differentially expressed genes" was accepted by Bioinformatics.
-
Dec. 30, 2020: RAD v1.2 is released. RAD now supports both hypergeometric or binomial test for statistical inference.
-
Oct. 24, 2020: RAD v1.1 is released. RAD now supports user-defined extended distance. We also update the illustrative diagram in the introduction.
-
Jul. 10, 2020: RAD v1.0 is launched.
Example Data
List of Publications Used RAD
- Liu, W., J. Gallego-Bartolomé, Y. Zou, Z. Zhong, M. Wang, S. P. Wongpalee, J. Gardiner, S. Feng, P. H. Kuo and S. E. Jacobsen., 2021. Ectopic targeting of CG DNA methylation in Arabidopsis with the bacterial SssI methyltransferase. Nature Communications 12(1): 3130. doi: 10.1038/s41467-021-23346-y
- Pastor, W.A., Liu, W., Chen, D., Ho, J., Kim, R., Hunt, T.J., Lukianchikov, A., Liu, X., Polo, J.M., Jacobsen, S.E. & Clark, A.T. (2018) TFAP2C regulates transcription in human naive pluripotency by opening enhancers. Nature cell biology, 20, 553-564. doi: 10.1038/s41556-018-0089-0
- Harris, C.J., Scheibe, M., Wongpalee, S.P., Liu, W., Cornett, E.M., Vaughan, R.M., Li, X., Chen, W., Xue, Y., Zhong, Z., Yen, L., Barshop, W.D., Rayatpisheh, S., Gallego-Bartolome, J., Groth, M., Wang, Z., Wohlschlegel, J.A., Du, J., Rothbart, S.B., Butter, F. & Jacobsen, S.E. (2018) A DNA methylation reader complex that enhances gene transcription. Science, 362, 1182-1186. doi: 10.1126/science.aar7854
- Gallego-Bartolomé, J., Liu, W., Kuo, P.H., Feng, S., Ghoshal, B., Gardiner, J., Zhao, J.M., Park, S.Y., Chory, J. & Jacobsen, S.E. (2019) Co-targeting RNA Polymerases IV and V Promotes Efficient De Novo DNA Methylation in Arabidopsis. Cell, 176, 1068-1082.e19. doi: 10.1016/j.cell.2019.01.029